Tumours are complex mixture of a heterogeneous population of cells that possess distinct genetic, morphological, and phenotypic profiles, which can affect tumour development and therapeutic resistance. The success of cancer treatments relies on the comprehension of the heterogeneous and immune hostile tumour microenvironment (TME), further on providing additional possibilities to eliminate its tumour supportive role.
Commonly used analytical methods give us little or no information about cell and biomarker spatial localization within tumour and their cross-talk during cancer treatments. Exploration of the diversity of cells has long been hampered by technical limitations. Immunohistochemistry traditionally allows staining of only a few markers at a time, while methods that are able to characterise the cells in great detail, such as single-cell sequencing and multiplex cytometry/CyTOF, are dependent on dissociation of the tissue, thereby losing the spatial context. To gain a full understanding of the TME, it is critical that we obtain a detailed picture of the phenotypes of cancer cells and activation state of immune cells, as well as their spatial organisation in the tissues including local and distal interactions; in other words their behaviour as a cellular system.
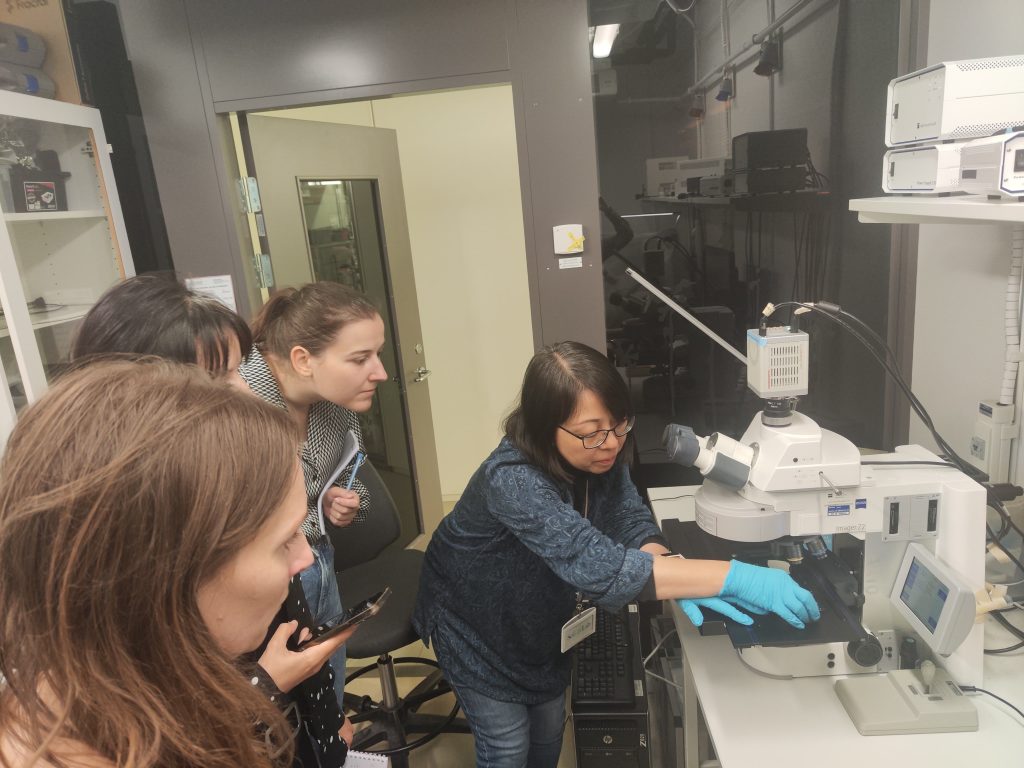
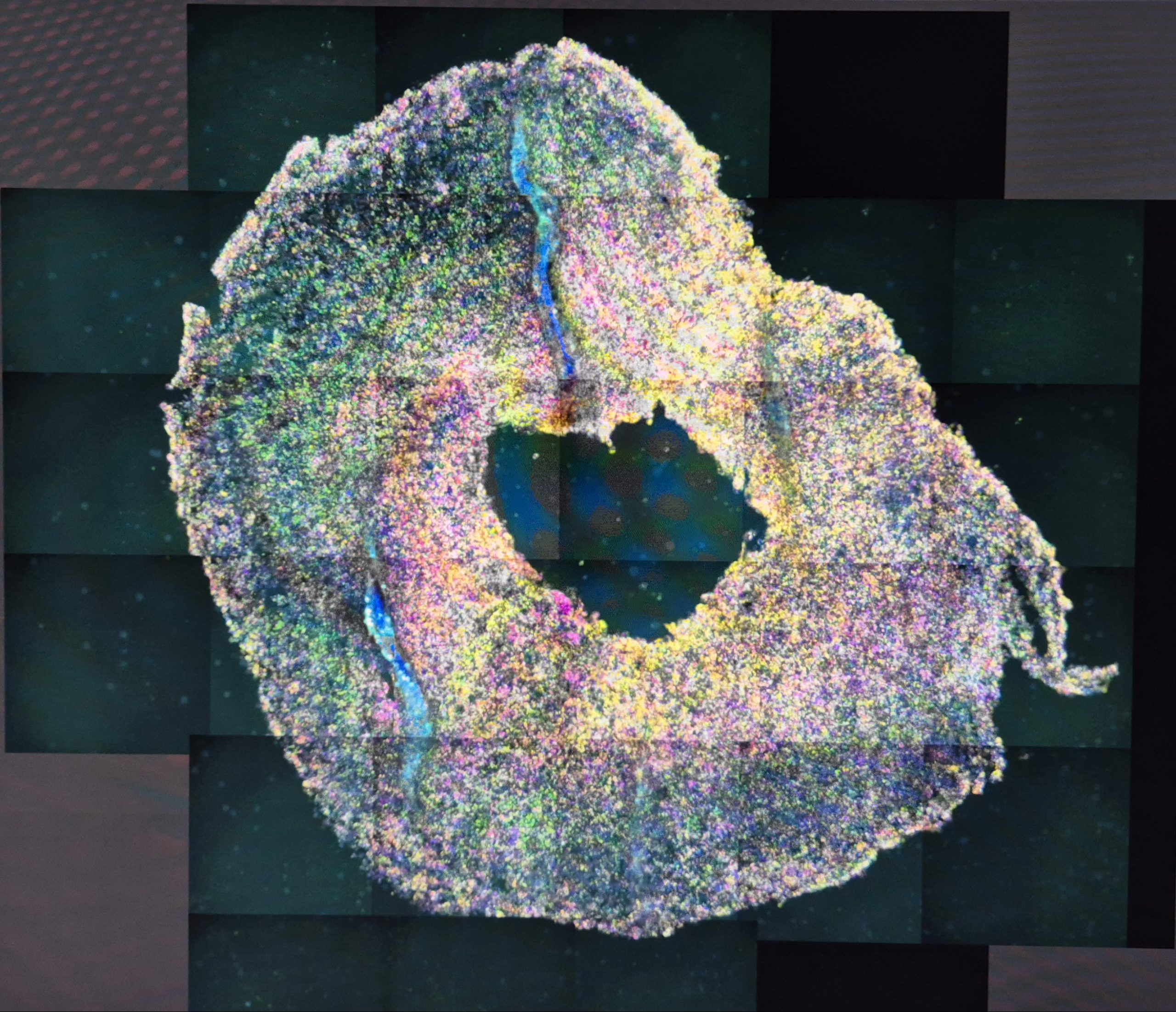
To achieve that, Mats Nilsson with his team at SciLifeLab in Stockholm organized a summer schools in ISS “In situ sequencing” profiling of brain tumours. ISS enables spatial and single-cell analyses of 3D cancer models and patient tumour tissues to characterize TME and to study the response of tumours to conventional and novel treatments, including radio-chemotherapy and immunotherapy, respectively. In situ sequencing (ISS) method for parallel targeted analysis of short RNA fragments of point mutations and multiplexed gene expression profiling in morphologically preserved cells and tissues of breast, lung and colon cancers, was first introduced in 2013 by the group of Mats Nilsson from SU. Improved ISS method called the hybridization-based in situ sequencing method (HybISS) was successfully demonstrated on human brain tissue samples, which are very challenging samples to study with image-based spatial analysis techniques. This also opens the door for the analyses of GBM and lung cancer samples. The week of Summer school combined both, lectures on ISS introduction and protocols by Mats Nilsson and Chika Yokota, lecture on Xenium presentation and demonstration by Katarina Tiklova, padlock probe design by Marco Grillo and data analysis lecture by Sergio Marco Salas, as well as hands-on training in the lab supervised by Maria Escriva Conde and Christina Bekiari. ISS methodology accomplished.